 |
 |
| “Give me alllll of youuuuu” - Pandas + hacky multiprocessing | “ill be done when I’m done” - Pandas |
TL;DR - Instead of doing df.apply(function), use the following code…
cores = mp.cpu_count()
partitions = cores
def parallelise(data, func):
data_split = np.array_split(data, partitions)
pool = Pool(cores)
data = pd.concat(pool.map(func, data_split))
pool.close()
pool.join()
return data
def wrapper(rows):
def function(row):
#do the expensive stuff
return #result
#df.apply syntax
return rows.apply(function)
_ = parallelise(df, wrapper)
Be weary of mindlessly using df.apply()
Say you have a function that you need to apply for every row in a pandas data frame.
Typically, if the function cannot be vectorised or done using pandas functions, one would resort to df.apply().
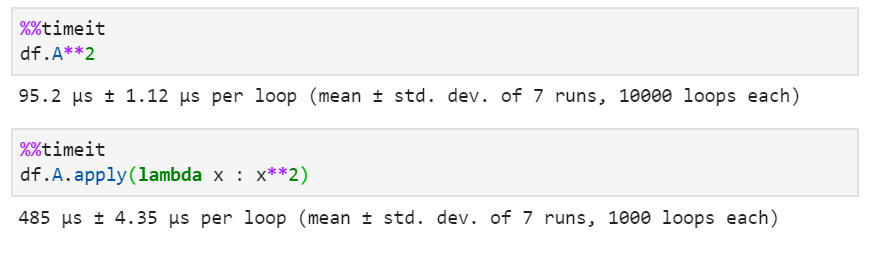
For the uninitiated, df.apply(f), takes a function f and applies along a particular axis of the data frame, df. For example, if you want to square every element in column A of the data frame df, you would do df.A.apply(lambda x: x**2). Although this is a terrible way to square number and should never be done in practice. Case in point, using df.A.apply() takes 5-times longer to do the same computation, and the code is much less readable than using df.A**2
What with these expensive functions?
OK, so… assuming that you have no other option than to use df.apply() for this operation, how can we speed it up?
Lets start with a concrete example. Say you have a data frame with 2 columns, A and B, that hold paths to images. We want to convert the images at these paths to HSV, calculate the mean HSV values for each image and compute the difference between the mean values of the image at A and the image at B. The code below outlines this operation. (rgb2hsv and imread from skimage)
def someExpensiveFunction(row):
imgA_mean = np.mean(rgb2hsv(imread(row.A)),axis=(0,1))
imgB_mean = np.mean(rgb2hsv(imread(row.B)),axis=(0,1))
hsv_mean_diff = imgA_mean - imgB_mean
return hsv_mean_diff
If i were to do vanilla df.apply, it takes 1min 2s to run for a data frame with about 600 image pairs (590 to be exact).
%%time
_ = df.apply(someExpensiveFunction,axis=1)
### Wall time: 1min 2s
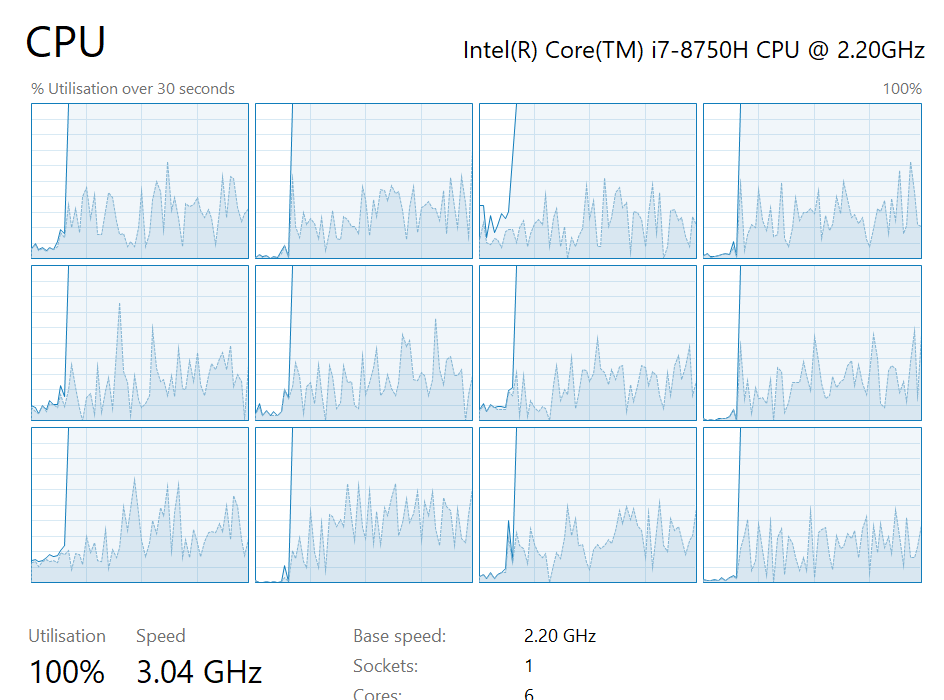
Currently this is barely using a single core on my PC so lets parallelise it.
Parallellllll
The parallelisation uses python’s built-in multiprocessing module, and is composed of 2 parts; the parallelise function and the worker function.
The worker function defines the actual calculation that needs to run and is quite similar to someExpensiveFunction. But, for this to work, we need to put someExpensiveFunction inside another function, which i will refer to as the wrapper function. This is someExpensiveFunction_wrapper in the code below. The wrapper function receives a block of data and applies the worker function to that data. The return statement of the wrapper function defines how this is done.
Think of rows as a smaller data frame, and you are applying someExpensiveFunction to that1.
def someExpensiveFunction_wrapper(rows):
def someExpensiveFunction(row):
imgA_mean =np.mean(rgb2hsv(imread(row.A)),axis=(0,1))
imgB_mean =np.mean(rgb2hsv(imread(row.B)),axis=(0,1))
hsv_mean_diff = imgA_mean - imgB_mean
return hsv_mean_diff
return rows.apply(someExpensiveFunction,axis=1)
The parallelise function splits the data frame into blocks and assigns each core to run wrapper function, on each data block.
cores = multiprocessing.cpu_count()
partitions = cores
def parallelise(data, func):
data_split = np.array_split(data, partitions)
pool = Pool(cores)
data = pd.concat(pool.map(func, data_split))
pool.close()
pool.join()
return data
And now, you invoke the parallelisation as follows
%%time
_ = parallelise(df, someExpensiveFunction_wrapper)
### Wall time: 22.3 s
The parallelised code ran about 3x faster than the original with much heavier use of the CPU.
Quick note for working in Jupyter notebooks on windows - To get this to work, I’ve had to copy
someExpensiveFunction_wrapperto an external python file and import into my Jupyter notebook. Structure is the same as before but now I have to doparallelise(df, extFile.someExpensiveFunction_wrapper)instead ofparallelise(df, someExpensiveFunction_wrapper). Not sure if this is a Jupyter issue or an issue with multiprocessing on windows.
So the complete code looks something like this…
Original
def someExpensiveFunction(row):
imgA_mean =np.mean(rgb2hsv(imread(row.A)),axis=(0,1))
imgB_mean =np.mean(rgb2hsv(imread(row.B)),axis=(0,1))
hsv_mean_diff = imgA_mean - imgB_mean
return hsv_mean_diff
_ = df.apply(someExpensiveFunction,axis=1)
### Wall time: 1min 2s
Modified
cores = mp.cpu_count()
partitions = cores
def parallelise(data, func):
data_split = np.array_split(data, partitions)
pool = Pool(cores)
data = pd.concat(pool.map(func, data_split))
pool.close()
pool.join()
return data
def someExpensiveFunction_wrapper(rows):
def someExpensiveFunction(row):
imgA_mean =np.mean(rgb2hsv(imread(row.A)),axis=(0,1))
imgB_mean =np.mean(rgb2hsv(imread(row.B)),axis=(0,1))
hsv_mean_diff = imgA_mean - imgB_mean
return hsv_mean_diff
return rows.apply(someExpensiveFunction,axis=1)
_ = parallelise(df, someExpensiveFunction_wrapper)
### Wall time: 22.3 s
And finally as code prototype to be copied…
cores = mp.cpu_count()
partitions = cores
def parallelise(data, func):
data_split = np.array_split(data, partitions)
pool = Pool(cores)
data = pd.concat(pool.map(func, data_split))
pool.close()
pool.join()
return data
def wrapper(rows):
def function(row):
#do the expensive stuff
return #result
#df.apply syntax
return rows.apply(function)
_ = parallelise(df, wrapper)
Be weary of mindlessly using Multiprocessing for everything
Although the parallelised code ran much faster than the original, there multiprocessing overhead needs to be accounted for. Spawning processes is expensive.
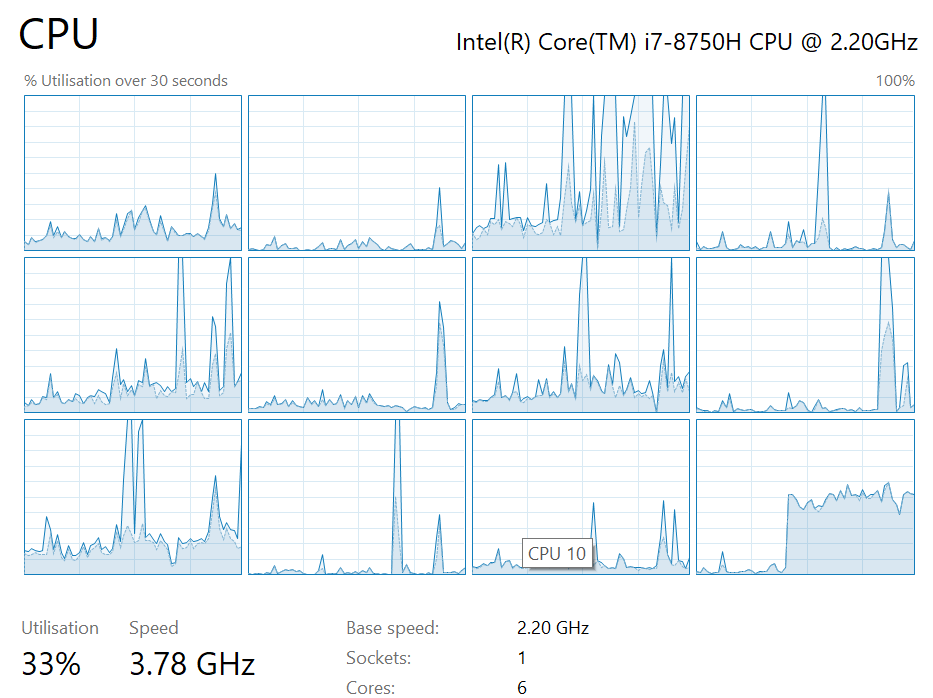
Consequently, for less expensive worker functions, parallelising would actually make the code run slower.
Parallelising the squaring example for before, using this approach made it about 3-times slower.
Yes… df.apply() with multiprocessing was slower than vanilla df.apply(). Given that was already 5x than df.A **2, the using multiprocessing would make it 15x slower.
On top of that, if you are running this on a laptop, chances are the sustained CPU clock for multiprocessing loads is lower compared to for single core load, due to thermal and power limits. So, as might be expected, performance is worse than simply multiplying the single thread time by the number of threads. Better performance could potentially be achieved on PC with a larger headroom to the TDP.
In general, the threshold computational cost of the worker function for multiprocessing to be worthwhile will vary depending on your hardware.
Off to the races!
Not really though… Don’t gamble kids… being fiscally responsible is cool!
 |
 |
| “I am Speed!” | Pandas playing where’s wally |
-
I know this looks like a job for decorators… ↩
